1. Graph Basics¶
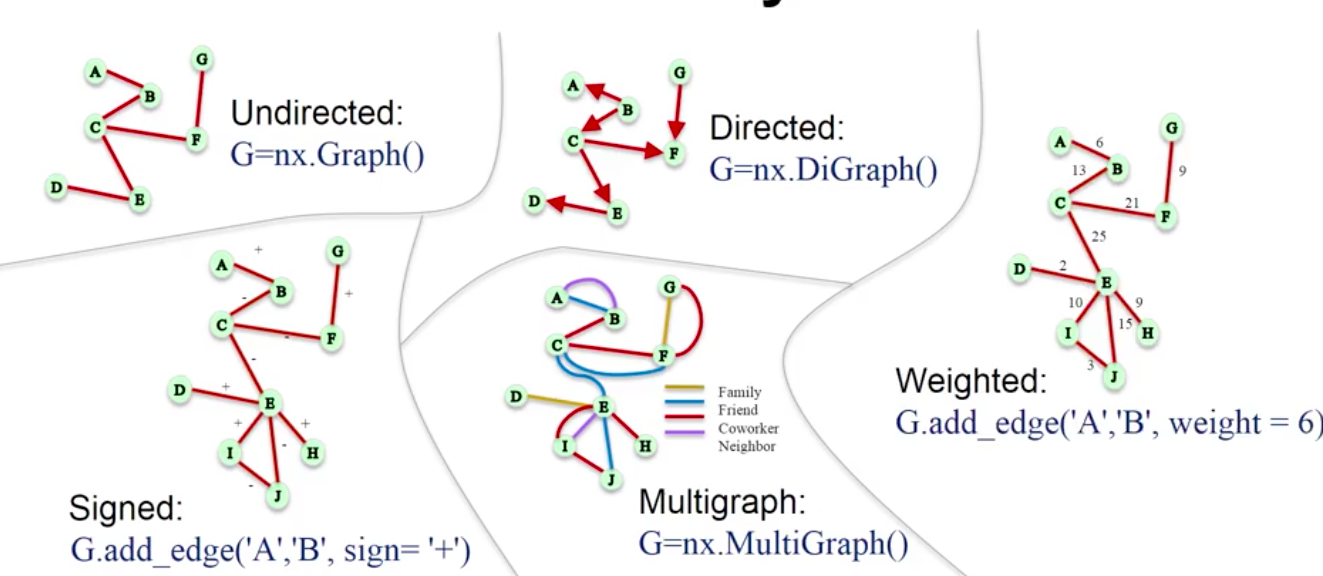
1.1. Basic Graphs¶
# Undirected Graph
G = nx.Graph()
G.add_edge('A','B')
G.add_edge('B','C')
# Directed Graph
D = nx.DiGraph()
D.add_edge('B','A')
D.add_edge('B','C')
# Check
D.is_directed()
# True
# Multi-Graph
M = nx.MultiGraph()
M.add_edge('B','A')
M.add_edge('B','C')
# Check
M.is_multigraph()
# True
# Directed Multi-Graph
DM = nx.MultiDiGraph()
DM.add_edge('B','A')
DM.add_edge('B','C')
# Check
DM.is_multigraph()
# True
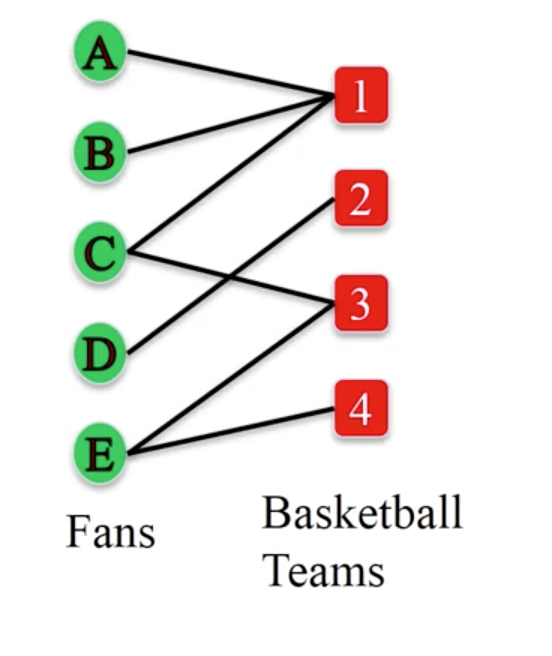
1.2. Bipartite Graph¶
Bipartite graph is a special network where there are two set of nodes, and nodes within each set have no edges between each other.
To create a bipartite graph:
from networkx.algorithms import bipartite
B = nx.Graph()
# Add nodes
B.add_nodes_from(['A','B','C','D','E'], bipartite=0)
B.add_nodes_from([1,2,3,4], bipartite=1)
# Add edges
B.add_edges_from([('A',1),('B',1),('C',1),('C',3),('D',2),('E',3),('E',4)])
# check if graph is bipartite
bipartite.is_bipartite(B)
# True
To get list of nodes with partitions
# get set of partition nodes
bipartite.sets(B)
To check if a set of nodes is part of a bipartition of a graph:
x = set([1,2,3,4])
print bipartite.is_bipartite_node_set(B, x)
# True
z = set([1,2,3,4,'A'])
print bipartite.is_bipartite_node_set(B, z)
# False
1.2.1. Projected Bipartite Graph¶
For each partition of a Bipartite Graph, it is possible to generate a projected graph where one set of nodes have common edges to the other set of nodes.
B = nx.Graph()
B.add_edges_from([('A',1), ('B',1), ('C',1),('D',1),('H',1), \
('B', 2), ('C', 2), ('D', 2),('E', 2), ('G', 2), ('E', 3), \
('F', 3), ('H', 3), ('J', 3), ('E', 4), ('I', 4), ('J', 4) ])
# set of nodes to generate a projected graph from a partition
X = set(['A','B','C','D', 'E', 'F','G', 'H', 'I','J'])
P = bipartite.projected_graph(B, X)
1.2.2. Weighted Projected Bipartite Graph¶
It is also possible to get the the weights of the projected graph using the function below.
bipartite.weighted_projected_graph(B, X)
1.3. Edge Types¶
# Weighted Edges
W = nx.Graph()
W.add_edge('A','B', weight=5)
W.add_edge('B','C', weight=6)
# Signed Edges
S = nx.Graph()
S.add_edge('A','B', sign='+')
S.add_edge('B','C', sign='-')
We can add edge attributes with any keys.
# Edge Attributes
R = nx.Graph()
R.add_edge('A','B', relation='friend')
R.add_edge('B','C', relation='coworker')
R.add_edge('B','D', relation='family')
And even add both weights & attributes
R.add_edge('A','B', relation='friend', weight=5)
1.4. Node Attributes¶
Same as edge attributes, nodes attributes can also be assigned with any keys.
G=nx.MultiGraph()
G.add_node('A',role='manager')
G.node['A']['role'] = 'team member'
G.node['B']['role'] = 'engineer'
1.5. Joining Two Graphs¶
Networkx can merge two graphs together with their differing weights when the edge list are the same.
new = nx.compose(a, b)
name1 name2 weights
Georgia Lee {u'Weight': 10}
Georgia Claude {u'weight': 3,u'Weight': 90}
Georgia Andy {u'weight': 1, u'Weight': -10}
Georgia Pablo {u'Weight': 0}
Georgia Frida {u'Weight': 0}
Georgia Vincent {u'Weight': 0}
Georgia Joan {u'Weight': 0}
Lee Claude {u'Weight': 0}