4. Connectivity¶
4.1. Clustering¶
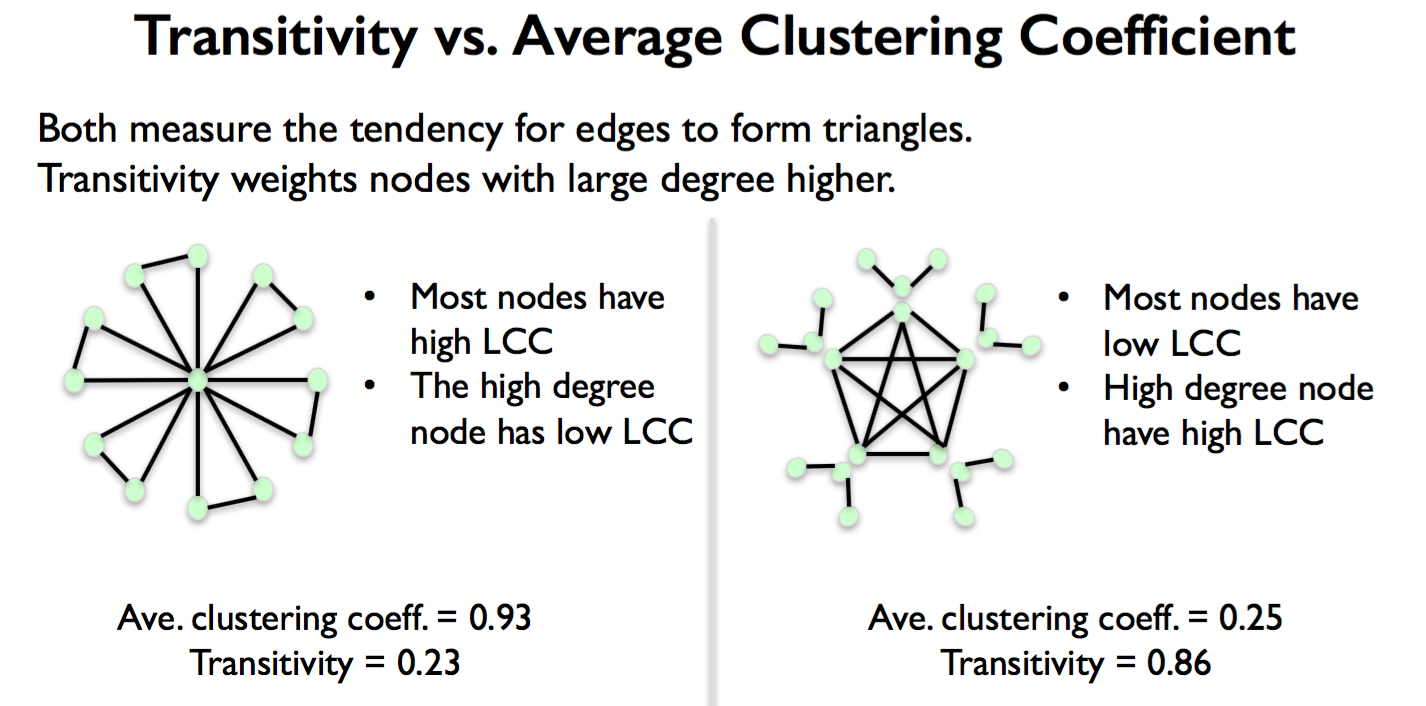
Clustering coefficients measures the degree to which nodes in a network tend to cluster or form triangles.
4.1.1. Local Clustering Coefficient¶
Find the clustering of a particular node.
# pairs of A’s friends who are friends with each other / # all possible pairs of A’s friends
# G is a graph, A is a node
nx.clustering(G, 'A')
4.1.2. Global Clustering Coefficient¶
Average clustering of entire network by averaging all local clustering coefficient values of all nodes.
nx.average_clustering(G)
Transitivity is the ratio of number of triangles and number of “open triads”. Puts larger weight on high degree nodes.
nx.transitivity(G)
4.2. Distance¶
4.2.1. Shortest Path¶
Shortest distance from a start node to the end node.
nx.shortest_path(G, 'A', 'H')
# ['A', 'B', 'C', 'E', 'H']
nx.shortest_path_length(G, 'A', 'H')
# 4
4.2.2. Longest Path¶
Eccentricity of a node n is the largest distance between n and all other nodes.
nx.eccentricity(G)
# {'A': 5, 'B': 4, 'C': 3, 'D': 4, 'E': 3, 'F': 3, 'G': 4, 'H': 4, 'I': 4, 'J': 5, 'K': 5}
Diameter maximum distance between any pair of nodes.
nx.diameter(G)
# 5
Radius of a graph is the minimum eccentricity.
nx.radius(G)
# 3
Periphery of a graph is the set of nodes that have eccentricity equal to the diameter.
nx.periphery(G)
# ['A', 'K', 'J']
Center of a graph is the set of nodes that have eccentricity equal to the radius.
nx.center(G)
# ['C', 'E', 'F']
4.2.3. Breadth First Search¶
Find the distance from one node to all other nodes.
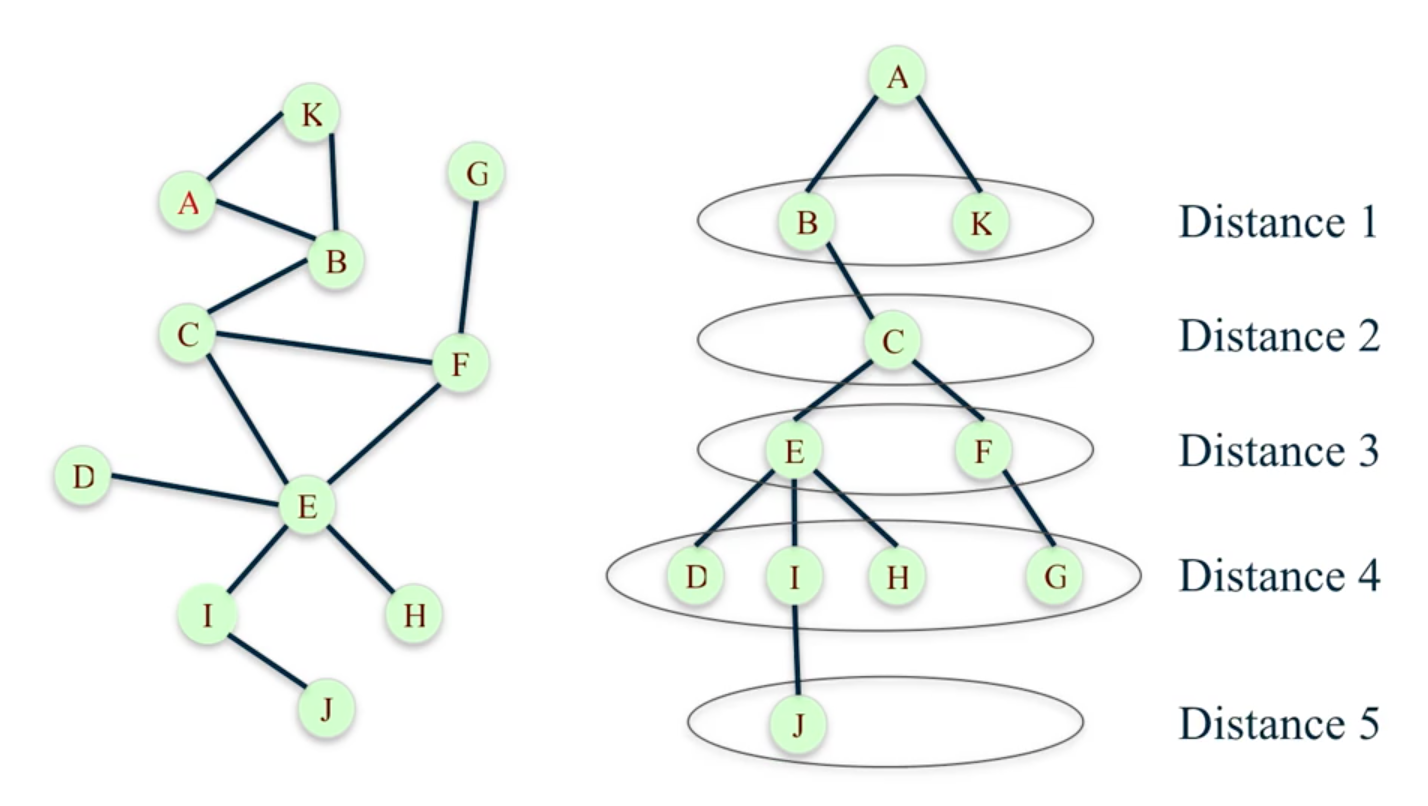
One method is the Breadth First Search, which is a systematic and efficient procedure for computing distances from a node to all other nodes in a large network by “discovering” nodes in layers.
T = nx.bfs_tree(G, 'A')
T.edges()
# [('A', 'K'), ('A', 'B'), ('B', 'C'), ('C', 'E'), ('C', 'F'),
# ('E', 'I'), ('E', 'H'), ('E', 'D'), ('F', 'G'), ('I', 'J')]
nx.shortest_path_length(G, 'A')
# {'A': 0, 'B': 1, 'C': 2, 'D': 4, 'E': 3, 'F': 3, 'G': 4, 'H': 4, 'I': 4, 'J': 5, 'K': 1}
4.2.4. Other Distance Measures¶
Average Distance between every pair of nodes.
nx.average_shortest_path_length(G)
# 2.52727272727
4.3. Connectivity¶
4.3.1. Unidirected Graph¶
Connected
An undirected graph is connected if, for every pair nodes, there is a path between them.
nx.is_connected(G)
Graph Components
To show nodes for each graph component.
# show all nodes for each components
sorted(nx.connected_components(G))
# show all nodes in component containing 'M'
nx.node_connected_component(G, 'M')
4.3.2. Directed Graph¶
Strongly / Weakly Connected
A directed graph is strongly connected if, for every pair nodes u and v, there is a directed path from u to v and a directed path from v to u.
nx.is_strongly_connected(G)
A directed graph is weakly connected if replacing all directed edges with undirected edges produces a connected undirected graph.
nx.is_weakly_connected(G)
Graph Components
A strongly connected graph component (subset of nodes) have (1) every node in the subset has a directed path to every other node. (2) no other node has a directed path to every node in the subset.
sorted(nx.strongly_connected_components(G))
# [{M}, {L}, {K}, {A, B, C, D, E, F, G, J, N, O}, {H, I}]
4.4. Network Robustness¶
Network robustness the ability of a network to maintain its general structural properties (connectivity) when it faces failures or attacks (removal of nodes or edges).
4.4.1. Disconnect a Graph¶
Disconnect by Node
What is the smallest number of nodes that can be removed from this graph in order to disconnect it?
nx.node_connectivity(G_un)
.. 1
# Which node?
nx.minimum_node_cut(G_un)
.. {'A'}
# can also choose source & target
nx.minimum_node_cut(G, 3, 7)
Disconnect by Edge
What is the smallest number of edges that can be removed from this graph in order to disconnect it?
nx.edge_connectivity(G_un)
.. 2
# Which edges?
nx.minimum_edge_cut(G_un)
.. {('A', 'G'), ('O', 'J')}
4.4.2. Disconnect Path¶
Imagine node G wants to send a message to node L by passing it along to other nodes in this network.
sorted(nx.all_simple_paths(G, 'G', 'L'))
# [['G', 'A', 'N', 'L'],
# ['G', 'A', 'N', 'O', 'K', 'L'],
# ['G', 'A', 'N', 'O', 'L'],
# ['G', 'J', 'O', 'K', 'L'], ['G', 'J', 'O', 'L']]
Disconnect by Node
If we wanted to block the message from G to L by removing nodes from the network, how many nodes would we need to remove?
nx.node_connectivity(G, 'G', 'L')
.. 2
# Which nodes?
nx.minimum_node_cut(G, 'G', 'L')
.. {'N', 'O'}
Disconnect by Edge
If we wanted to block the message from G to L by removing edges from the network, how many edges would we need to remove?
nx.edge_connectivity(G, 'G', 'L')
.. 2
# Which edges?
nx.minimum_edge_cut(G, 'G', 'L')
.. {('A', 'N'), ('J', 'O')}